March 2018
Ryegrass is the predominant grass weed species occurring in the small grain production areas of the Western Cape. Ryegrass is genetically highly variable and can cross-pollinate over some distance. This weed can rapidly develop herbicide resistance, especially to herbicides from Group A (fops, dims and den) and Group B (SU’s, IMI’s and TP’s).
The occurrence of herbicide resistance has had a negative impact on the control options of this weed. Making use of DNA markers to identify resistance has proven to be a quick method for in-season screening for target-site resistance. Since molecular genotyping makes use of the plants’ DNA, it is also diagnostically very accurate.
What is target-site herbicide resistance?
Herbicide resistance can be defined by the inherent ability of a weed to survive an herbicide application that would otherwise kill a normal plant/biotype/population of the same weed species. In any given weed population, there will be a plant that is genetically resistant to the applied herbicide. There are two main ways that a weed can develop resistance to herbicides.
Target-site resistance occurs when a change takes place in the protein that binds the herbicide. An example would be where a mutation within the target protein reduces or eliminates binding of the herbicide. It is very common to get cross-resistance to other herbicides of the same herbicide mode of action. Target-site resistance commonly involves herbicides from Group A and Group B.
Non-target site resistance most commonly occurs due to an increased herbicide detoxification process. Enhanced metabolism is also typically observed in herbicides that are used selectively, such as Group A and Group B. Reduced translocation also falls under this resistance group.
Research results 2015 - 2017
Mutation markers can be used to identity target-site mutations in weed species. These markers are then used to genotype specific resistant biotypes for specific mutations. Since 2013, ARC-Small Grain has been working on optimising the markers for South African ryegrass.
Through the screening of multiple samples, two very distinct mutations have been identified in South African ryegrass populations occurring in small grain producing areas.
These markers are the ALS mutation markers 197, which confers resistance to all group B herbicides (SU’s, Triazolopyrimidines and Imidazolinones) and ACCase mutation 2078, which confers resistance to many Aryloxyphenoxy-propionates, all Cyclohexanediones (including clethodim) and Phenylpyrazolines.
This means that all grass weed samples that are received, are screened for these mutations first, but if none of the mutations are present in the sample, the sample is screened with all other mutation markers. This is done to make sure that no mutation is missed in the screening process.
From 2015 to 2017, 165 samples from different fields/farms were received. At least four replicates were performed per sample, but often six to eight replicates were performed if sufficient sample material was received. The samples were all screened for various ACCase and ALS mutations. Resistance to more than one mutation were often recorded and most samples showed resistance to both ACCase and ALS mutations.
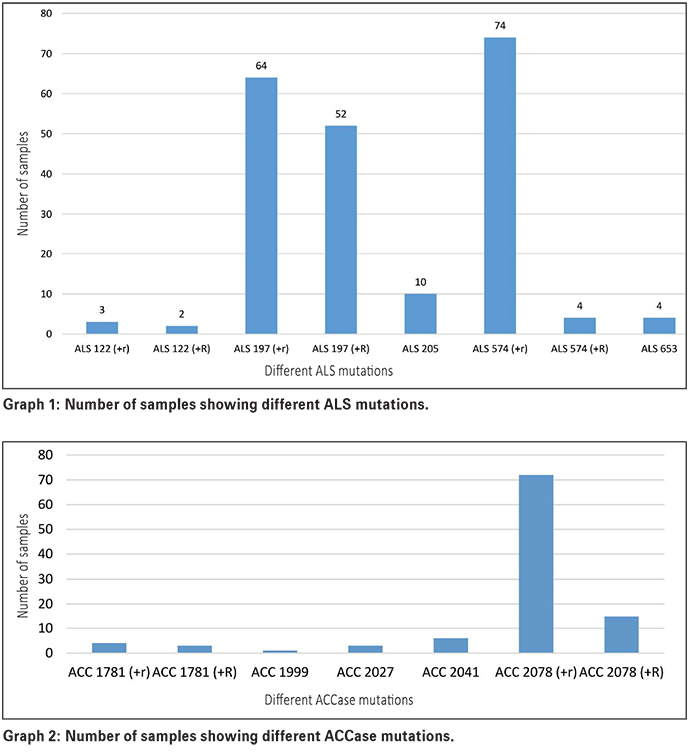
Graph 1 indicates the number of samples that showed resistance to the various ALS mutations. The mutation markers also distinguished between plants that are heterozygous (+r) (one copy – resistant allele) or homozygous (+R) (two copies – resistant allele) resistant for each specific target-site mutation. It is important to note that one sample could show heterozygous and homozygous resistant individuals, as at least four replicates were included per sample.
From the results, most samples showed heterozygous resistance to ALS mutation marker 574 (74), followed by heterozygous resistance to ALS mutation marker 197. ALS mutation markers 574 and 197 confer target-site resistance to IMI’s, SU’s and TP’s. What is worrisome, is that at least one replicate of 52 samples showed a homozygous resistant ALS 197 mutant. Very low numbers of the other ALS mutation markers were observed.
Graph 2 indicates the number of samples that showed resistance to the various ACCase mutations. The mutation markers again distinguished between plants that are heterozygous (+r) (one copy – resistant allele) or homozygous (+R) (two copies – resistant allele) resistant for each specific target-site mutation. It is again important to note that one sample could show heterozygous and homozygous resistant individuals, as at least four replicates were included per sample.
From the results, most samples showed heterozygous resistance to ACCase mutation marker 2078, followed by homozygous resistance to the same mutation marker. ACCase mutation marker 2078 confers resistance to many fops and all dims, including clethodim.
When heterozygous/homozygous genotypes to ACCase herbicides are detected, most ACCase herbicides including clethodim, clodinafop, diclofop, fluazifop, haloxyfop, butroxydim, sethoxydim, tralkoxydim and pinoxaden should be avoided.
Summary
From data obtained from screening 165 different samples over three years, it is evident that target-site resistance to ACCase- and ALS inhibitor herbicides is very common in South Africa. The most frequently observed target-site mutation was for ALS marker 197 and ACCase marker 2078.
These confer a broad cross-resistance in varying levels to most ACCase- and ALS herbicides. By knowing the mutations that occur in South Africa, a true resistance profile can be established for target-site resistant biotypes, allowing for more informed management choices.
 For further information regarding this topic, please contact Hestia Nienaber at 058 307 3420 or deweth@arc.agric.za or Scott Sydenham at sydenhams@arc.agric.za.
For further information regarding this topic, please contact Hestia Nienaber at 058 307 3420 or deweth@arc.agric.za or Scott Sydenham at sydenhams@arc.agric.za.

Publication: March 2018
Section: Focus on